Recent progresses in DNA nanotechnology have allowed the accomplishment of an almost infinite variety of nanosized objects of the most appealing shapes, endowed with hundreds of precisely located specific reaction sites. Can these DNA objects be exploited as ordering scaffolds to selforganize matter at the nano-scale? It was proven that different protein molecules can be arranged at specific positions on DNA scaffolds in solution, but then the problem arises of how to arrange and connect the nanoscaffolds on a wider, usable substrate, again in an ordered and addressable fashion.
A collaborative work between Centro NAST, ENEA, the cDNA Center of the University of Aarhus and the Center for Nanoscale Materials Science of the Oak Ridge National Laboratory, attempted the connection of such DNA nanoscaffolds to inorganic conductors (gold) orderly organized on a silicon dioxide surface; this aims at exploiting their potential as “nano-breadboards” for selfassembly of organic components and inorganic nanoparticles on them.
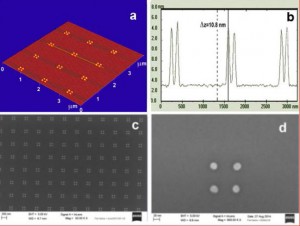
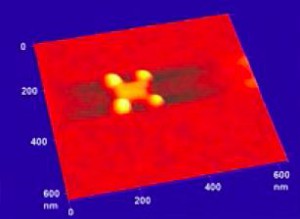
The team chose the simplest geometry of DNA nano-breadboard, a flat rectangular shape of approximately 75nm x 85nm, as thick as the DNA double helix, 2 nm. Even this simple and small board allows for bonding of more than 200 different components per face, on different specific locations on the boards, each determined within approximately 30 nm2. DNA boards are selfassembled from a long single stranded DNA represented by the M13 bacteriophage genome and 224 different oligonucleotides having selected sequences that hybridize different parts of the backbone and “staple” them together in the desired shape. This is the so called “DNA origami” technique. The staples at the corners of the rectangle are extended with sequences bearing two sulphur atoms that can later make covalent bonds to suitably arranged anchoring gold nanodots fabricated by electron beam lithography. And each of the stapling sequences can be extended with specifically designed sequences in order to bind the desired molecular components. Once the boards are assembled and purified they are allowed to react in solution with the gold nanodots, so that they can deposit and immobilize on them. In the first experiments, where groups of 4 nanodots at the corner of an 80 nm side square were used, the rate of correctly occupied nanodots was approximately 10%, which is satisfactory considering the statistical character of the origami-nanoanchor encounter, and their small size; nanodots are about 20 nm in diameter and 7-8 nm tall. Development of dielectrophoresis techniques to increase the rate of reaction is underway. Figure 1 shows AFM and SEM characterization of the gold nanodots, while Figure 2 shows an AFM topographic image of a DNA origami nanobreadboard docked and suspended onto its four gold nanodots. These will be later connected to an electrical detection system.
Authors from NAST Centre: Piero Morales, Mario Caruso, Liqian Wang, Antonio Rinaldi, Stefano Vespucci
Other Contributors: Kurt Gothelf, Mattia De Stefano, Abhichart Krissanaprasit, Jesper Vinther (Aarhus University); Claudia Dalmastri, Lucia Mosiello (ENEA), Scott Retterer (CNMS Oak Ridge Nat. Lab.)
Related Links and Media


