Nanotechnology is the science of manipulating matter at the atomic and molecular level to obtain materials with specifically enhanced chemical and physical properties. Collaborative research among scientists from Italy – the Nast Centre for Nanoscience Nanotechnology and Innovative Instrumentation, are working in the emerging challanging field of nanomedicine (the preservation and improvement of human health using the tools and knowledge of nanotechnology) may permit the development of more effective means for diagnosing and treating malignancies compared to the currently used small molecules. To enhance diagnostic or therapeutic efficacy, novel nanoassemblies must be engineered to function in biologically relevant environments and have multivalent loading capacity for easy detection or effective drug delivery. In the past decade, several types of nanoparticles have been prepared and evaluated for tissue targeting, sensing and imaging, and localized therapy (Akerman et al., 2002, Bottini et al., 2006). Often, the chemistry of these nanoparticles has limited their stepwise assembly into multilayered systems and capacity to interact multivalently with cell membrane receptors. Silica nanoparticles (SNP) have been widely used for biosensing and catalytic applications due to their large surface area to volume ratio, straightforward manufacture and the possibility to be doped and/or functionalized with fluorescent molecules (Bottini et al., 2006), magnetic nanoparticles (Wang et al., 2005) or semiconducting nanocrystals (Lin et al., 2006). SNP can be prepared using several techniques, including the widely used Stobër technique and the water-in-oil nanoemulsion system. The former is based on the simple hydrolysis of a silica precursor in alcoholic medium in the presence of ammonia; the latter uses water droplets inside of reverse micelles as nanoreactors. The size of the final nanospheres is mainly regulated by the dimension of the water droplets and, therefore, by the molar ratio of water to surfactant and by the molar ratio of water to the precursor. Other relevant parameters are the molar ratio of the precursor to the catalyst, the reactivity of the precursor, the total time and temperature of the reaction. Quantum dots (QD) are colloidal semiconducting nanocrystals a few nanometers in diameter, and are typically coated with functional polymers that permit the tethering of various small and macro-molecules resulting in aqueous environment compatibility. Because of their small size, their energy levels are quantized so that photon energy emission is restricted to a narrow range after broad range absorption. Their adherence to quantum behavior makes them superior to small organic fluorescent molecules that bleach rapidly after excitation only in a narrow absorption range. Therefore, QD represent an excellent building block for the realization of intracellular nanoprobes and delivery systems. However, QD have shown signs of toxicity dependent on several factors like dose, dimension and chemical functionalization (Hardman, 2006). Moreover their solubilization under physiological conditions and superficial funzionalization are not straightforward. Encapsulation of QD into SNP could increase their wettability and decrease the cytotoxic effects (Zhang et al., 2006). Moreover specific functional groups can be easily introduced on the silica surface reducing aggregation and allowing further functionalization to realize nanoengineered nanoassemblies (Bag-we et al., 2006). In this report we describe the realization and characterization of SNP doped with QD and decorated with neutravidin (Nav). To demonstrate that this luminescent nanoassembly could be further functionalized with either small or macromolecules, we coated the surface with biotilynated mouse anti-human CD3 (αCD3), exploiting the strong affinity between biotin and Nav, and we investigated their delivery into Jurkat T cells through CD3 receptor-mediated endocytosis. As such, the realized nanoassembly represents a potentially excellent scaffold for constructing intracellular multivalent nanoprobes.
MATERIALS AND METHODS
Materials
Unless otherwise noted, all reagent-grade chemicals were used as received, and Millipore water was used in the preparation of all aqueous solutions. Cyclohexane, Triton X-100, n-hexanol, tetramethyl orthosilicate (tmos), aminopropyl trimethoxysilane (apts), trihydroxysilyl propylmethylphosphonate (thpmp), ammonia (28% in water), poly-L-lysine, formaldehyde, dimethyl formamide (DMF), Triton X- 100 and PBS were purchased from Sigma-Aldrich (St. Louis, MO). ZnS-covered CdSe QDs from Evident Technologies, Inc. (Troy, NY) were capped by trioctylphosphine oxide (TOPO) had 542-nm emission wavelengths and diameter of approximately 2 nm. Water-soluble biotin labeling reagents with spacer arm (sulfo-NHS-LC-biotin) and Nav were purchased from Pierce Biotechnology, Inc. (Rockford, IL). Biotinylated and unconjugated αCD3 were from eBioscience, Inc. (San Diego, CA); rabbit anti-human CD107A (LAMP-1) antibody was received from Prof. Fukuda laboratory (Burnham Institute for Medical Research, La Jolla, CA). Texas Red-labeled goat anti-rabbit antibody and Texas Red-labeled concanavalin A were from Molecular Probes (Invitrogen Corp., Carlsbad, CA); normal mouse serum (NMS) was from Santa Cruz Biotechnology (Santa Cruz, CA); normal goat serum (NGS) was from Gibco (Invitrogen Corp., Carlsbad, CA); fetal bovine serum (FBS) was from Tissue Culture Biologicals (Informagen, Inc., Newington, NH); RPMI-1640 cell culture medium and Dulbecco’s modified Eagle’s medium-high glucose (DMEM-HG) were from Cellgro (Mediatech, Inc., Herndon, VA).
Realization quantum dot-doped silica nanoparticles decorated with neutravidin
A cyclohexane, Triton X-100 and n-hexanol mixture (volume ratio 4.2:1:1; the final volume of the mixture was 24.8 ml) was converted to a nanoemulsion by stirring at room temperature for 1h. Water (940 μl) and tmos (100.5 μl) were added. Sonication for 1 h was performed to facilitate the diffusion of tmos into the encapsulated water droplets in the nanoemulsion. A drop (59 μl) of ammonia was added to catalyze the hydrolysis of tmos and condensation.
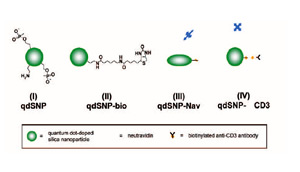
Scheme 1 – Quantum dot-doped silica nanoparticles (I) functionalized with amino and phosphonate groups, (II) after the coupling of the amino groups to biotin labeling reagents with spacer arm, (III) decorated with neutravidin and (IV) after incubation with biotilynated mouse anti-human CD3.
The mixture was stirred for 30 min, and then 25 nmoles of QD in toluene were added. The reaction mixture was stirred for 24 h, followed by the addition of tmos (10.05 μl), and a mixture of apts (5.9 μl) and thpmp (15.1 μl). The time interval between the addition of tmos and the mixture of organosilanes was at least 30 min. The mixture was further stirred for 24 h, and then the quantum dot-doped SNPs (qdSNP) were precipitated by addition of acetone (25 ml) and washed several times with water then anhydrous ethanol (scheme 1(I)). 1 mg of qdSNP were stirred for 2 hours in phosphate saline buffered (PBS) solution in presence of 30 mg of sulfo-NHS-LC-biotin and then washed 3 times with PBS (qdSNP-bio, scheme 1(II)). qdSNP-bio were incubated with 5 mg of neutravidin overnight and then washed with PBS until the absorbance of the collected supernatant revealed the total absence of free protein (qdSNP-Nav, scheme 1(III)).
Cell internalization by CD3-mediated endocytosis
qdSNP-Nav were incubated with biotinylated αCD3 antibody for 1 hour and washed one time with PBS (qdSNP-αCD3, scheme 1(IV)). Jurkat T leukemia cells were kept in logarithmic growth by culture in RPMI-1640 supplemented with 10% FBS. The cells were washed twice with PBS, resuspended in 2 ml of PBS and incubated for 1 h at 4 °C on poly- L-lysine-coated cover slips. HeLa adenocarcinoma cells were seeded onto cover slips and kept in logarithmic grown by culture in DMEM-HG for 24 h. The cover slips were treated with qdSNP-αCD3 in PBS for 4 h at 37 °C in 5% CO2 atmosphere, fixed for 10 min in 3.7% formaldehyde in PBS, and then either stained for 1 h at room temperature with 10 μg/mL Texas Red-conjugated concanavalin A or incubated with rabbit anti-CD107A and then with Texas Redlabeled goat anti-rabbit antibody in PBS containing 5% NGS and 0.1% Triton X-100. VECTASHIELD mounting medium with DAPI (Vector Laboratories, Inc., Burlingame, CA) was used to visualize the nuclear region.
Fluorescent Microscopy
Phase-contrast and fluorescence images of functionalized SNPs were acquired using an Inverted TE300 Nikon (Kanagawa, Japan). Confocal microscopy (Radiance 2100/AGR-3Q, Bio Rad, Hercules, CA) was used to collect fluorescent images. Both 40x and 60x (1.4-oil immersion) objectives were used. To avoid fluorescence interference from salt crystals caused by evaporation of buffer, wet samples were imaged.
Trasmission Electron Microscopy
An aliquot of qdSNP in ethanol was dropped onto the lacey carbon film covering a 300-mesh copper
grid (Tedpella, Inc., Redding, CA) and let to evaporate. TEM images were obtained using a Hitachi H-600A (Tokyo, Japan).
RESULTS AND DISCUSSION
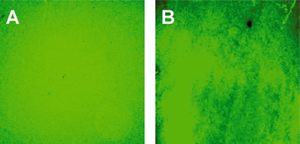
Realization of quantum dot-doped silica nanoparticles decorated with neutravidin TEM images of qdSNP showed uniform diameter (13±1 nm) silica nanobeads. The presence of phosponate groups facilitated the dispersion in PBS and the subsequent steps of coupling of the amino groups to biotin labeling reagents with spacer arm and of decoration with Nav. qdSNP-Nav in PBS remained as a clear solution without visible flocculation for several months, suggesting that the nanoassemblies existed as monodispersed suspension. The latter result was confirmed by the fact that both optical and confocal microscopy showed that qdSNP-Nav in PBS had no distinct fluorescent features (figure 1).
Figure 1 – Optical (A) and confocal (B) fluorescent images of quantum dot-doped silica nanoparticles decorated with neutravidin dispersed in PBS. That both fields displayed uniform fluorescence suggests that the nanoassemblies were fully dispersed in PBS (scale bar approximately 10 μm).
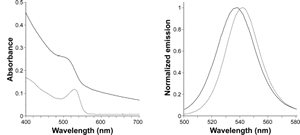
Absorbance spectrum of qdSNP-Nav dispersed in PBS showed a band which was broader and blue-shifted compared to the corresponding 531 nm-centered band in the spectrum of QD dissolved in toluene (figure 2A).
Figure 2. – Absorbance in the visible range (A, far left) and normalized emission (B, left) spectra of quantum dots in toluene (solid line) and quantum dot-doped silica nanoparticles decorated with neutravidin dispersed in PBS (dotted line).
On excitation at 360 nm, the steady-state emission spectrum of qdSNP-Nav in PBS exhibited slightly broader and blue-shifted emission peak compared to that of free QD in toluene (figure 2B and table 1). These results suggest that the encapsulation into the silica host did not affect the emission properties of the semiconducting nanocrystals. Moreover the decoration with neutravidin permitted to stably dissolve the nanoassemblies under physiological conditions and therefore to obtain a potential scaffold for intracellular transporters. Cell internalization by CD3-mediated endocytosis Recently, the transporting of various types of proteins non-covalently and nonspecifically bound to nanoengineered nanomaterials through endocytosis into various adherent and nonadherent mammalian cell lines was reported.
| Sample |
λmax† (nm) | FWHM‡ (nm) |
| QD in toluene | 542 | 28 |
| qdSNP-Nav in PBS | 538 | 32 |
Table 1 – Spectroscopic parameters, maximum emission wavelength and full-width half-maximum, for dipersions of QD in toluene and qdSNP-Nav in PBS (*maximum emission wavelength; ** full-width half-maximum).
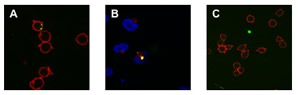
We investigated whether qd- SNP-Nav could be used as fluorescently detectable intracellular nanoprobes in Jurkat cells through specific CD3 receptor-mediated endocytosis. qdSNPNav was preliminarly incubated with biotilynated αCD3 to obtain SNP functionalized with αCD3 (qd-SNP-αCD3). Endocytotic uptake pathways are energy- dependent, with optimum uptake occurring at 37ºC and none occurring at 4ºC. Jurkat cells that had been incubated with qdSNP-αCD3 at 37ºC had intense internal fluorescence (figure 3A).
Figure 3 – A: Confocal fluorescent images of qdSNP-Nav internalized into Jurkat T leukemia cells through biotinylated αCD3-CD3 receptor- mediated endocytosis. B: Confocal fluorescent images of qdSNP-Nav internalized into Jurkat T leukemia cells incubated with rabbit anti-CD107A and then with Texas Red-labeled goat anti-rabbit antibody in order to visualize lysosomes. The co-localization of intracellular nanoassemblies (green) and lysosomes (red) indicate that after CD3 receptor-mediated endocytotic uptake, part of internalized qdSNP-Nav were transported to lysosomes. C: Confocal fluorescent images of Jurkat cells treated at 37ºC with SNP-Nav previously incubated with unconjugated αCD3.
Jurkat cells were treated with VECTASHIELD mounting medium with DAPI in order to visualize the nuclear region and to investigate if internalized qdSNP-αCD3 reached the nucleus. Intracellular nanoassembly did not overlap the DAPI stained region suggesting that the nanoassembly, after the endocytotic uptake, was not transported to the nucleus. Further, Jurkat cells were incubated with rabbit anti-CD107A and then with Texas Red-labeled goat anti-rabbit antibody in order to visualize the lysosomes, the organelles that serves to digest macromolecules through digestive enzymes. The presence of co-localized red and green features suggested that after the cellular uptake some endocytotic vesicles released the qdSNP-αCD3 to the lysosomes (figure 3B). Jurkat cells that had been treated with qdSNP-αCD3 at 4ºC or those treated with SNP-Nav previously incubated with unconjugated αCD3 (at 4ºC or 37ºC) exhibited weak internal fluorescence (figure 3C). Moreover, we incubated adherent HeLa cells with qdSNP- αCD3. Extremely weak internal red fluorescence was observed in few cells. These results suggest that without stimulation of the CD3 membrane receptor (qd- SNP-Nav treated with unconjugated αCD3) or in absence of the CD3 surface receptor (HeLa cells) the endocytotic uptake of the nanoassembly was weak, probably due to non-specific interactions between the nanoassembly and hydrophobic regions of the cell surface. On the other hand, we observed that the endocytotic uptake of the qdSNP-Nav was strongly amplified if it occurs by specific CD3 receptor-mediated endocytosis with the αCD3 acting as a bifunctional agent to crosslink the CD3 surface receptor to the nanoassembly through biotin-streptavidin binding. Further, we observed that, once internalized by cells, part of the nanoassembly was transported to lysosomes.
CONCLUSIONS AND FUTURE PROSPECTS
In conclusion we fabricated silica nanobeads doped with quantum dots and coated with neutravidin (qd- SNP-Nav). The resulting nanoassemblies were visible by conventional fluorescent microscope and could be further functionalized with macromolecules, nucleic acids and polymers exploiting the strong affinity between biotin and Nav. This realized luminescent nanoassembly was delivered internally into Jurkat T cells through biotinylated- αCD3-CD3 receptor-mediated endocytosis and was partially released to lysosomes. As such, qdSNPNav represents a potentially excellent scaffold for constructing intracellular multivalent nanoprobes. In the future we plan to use this nanoassembly to investigate whether a synthetic pH-responsive polymer can enhance intracellular trafficking to the cytoplasm or to deliver nucleic acids into a specific cell line.
Experimental Teams: Massimo Bottini, Federica D’Annibale, Eleonora Nicolai, Almerinda Di Venere, Giampiero Mei, Andrea Magrini, Antonio Bergamaschi, Nicola Rosato
References
M. E. Akerman, W. C. Chan, Laakkonen, S. N. Bhatia, E. Ruoslahti. Proc. Natl. Acad. Sci. USA 2002, 99: 12617-12621.
R. P. Bagwe, L. R. Hilliard, W. Tan. Langmuir 2006, 22: 4357-4362.
M. Bottini, F. Cerignoli, M. I. Dawson, A. Magrini, N. Rosato, T. Mustelin. Biomacromolecules 2006, 7: 2259-2263.
M. Bottini, A. Magrini, A. Di Venere, S. Bellucci, M. I. Dawson, N. Rosato, A. Bergamaschi, T. Mustelin. J. Nanosci. Nanotechnol. 2006, 6: 1381-1386.
R. Hardman. Environ. Health Perspect. 2006, 114: 165-172.
Y. W. Lin, C. W. Liu, H. T. Chang. J. Nanosci. Nanotechnol. 2006, 6: 1092-1100.
T. Zhang, J. L. Stilwell, D. Gerion, L. Ding, O. Elboudwarej, P. A. Cooke, J. W. Gray, A. P. Alivisatos, F. F. Chen. Nano Lett. 2006, 6: 800-808.
J. Wang, K. Zhang, Y. Zhu. J. Nanosci. Nanotechnol. 2005, 5: 772-775.
_______________________________________________________________________
Results from BIONANO PROJECT:
Smart Nanomaterials for Biomedical Applications
Participants: Nicola Rosato (Scientific Coordinator), Adalberto Balzarotti, Piero Chiaradia, Maurizio De Crescenzi, Rodolfo Del Sole, Almerinda Di Venere, Giampiero Mei, Pietro Morales, Gaio Paradossi, Fulvia Patella, Riccardo Polini, Wolfgang Richter, Roberto Senesi, Lorenzo Stella, Mariano Venanzi
Related Information
Prof. N. Rosato
Lipoxygenases